Neurips 2023 has been incredibly awesome to scan through. The paper list is long and behind paywall, but usually searching for the paper titles will bring something up in arxiv or some tweet-thread related to it.
Patrick Mineault (OG Building 8 ) has collected a list of NeuroAI papers from Neurips which has been very useful to scan through.
Quirky papers
Time Series as Images: Vision Transformer for Irregularly Sampled Time Series
Instead of trying to figure out how to align differently sampled time series for time series classification task, plot them and send the image form to vision transformer, add a linear prediction head on top and be done with it.
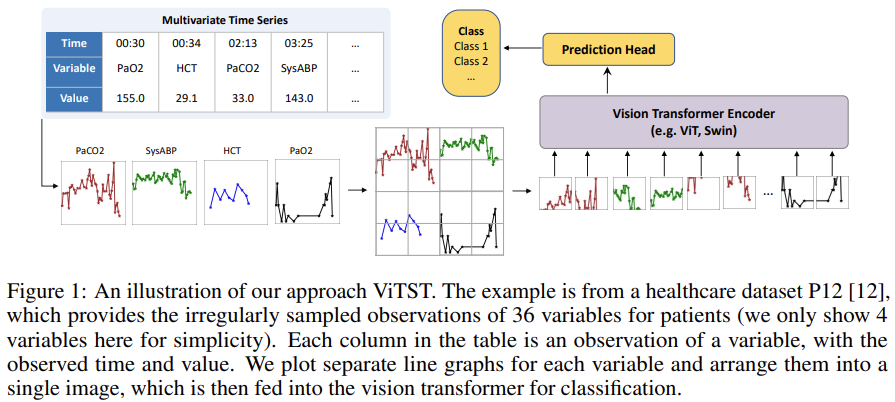
And this actually works:
We conduct a comprehensive investigation and validation of the proposed approach, ViTST, which has demonstrated its superior performance over state-of-the-art (SoTA) methods specifically designed for irregularly sampled time series. Specifically, ViTST exceeded prior SoTA by 2.2% and 0.7% in absolute AUROC points, and 1.3% and 2.9% in absolute AUPRC points for healthcare datasets P19 [29] and P12 [12], respectively. For the human activity dataset, PAM [28], we observed improvements of 7.3% in accuracy, 6.3% in precision, 6.2% in recall, and 6.7% in F1 score (absolute points) over existing SoTA methods.
Even though most of the “plot” image is simply empty space, attention map shows the transformer is attending the actual lines, and regions with more changes.
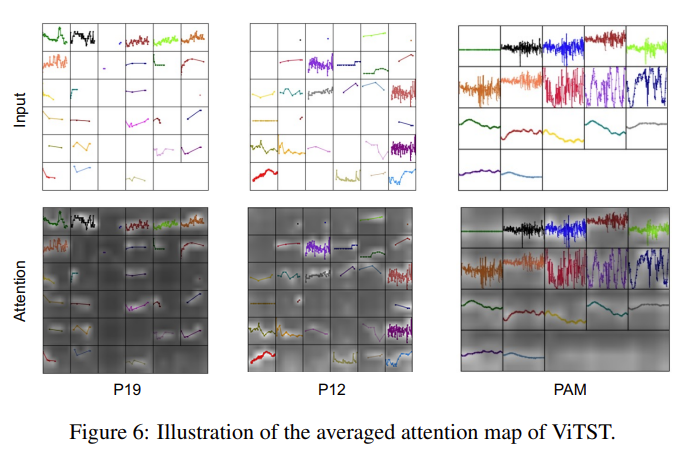
Why does this work? I’d think it’s because the ViTST acts as an excellent feature extractor, since the DL vision models contains in them representations of primitive features typically present in the line signals (e.g. edges, curves, etc). Yet using a pretrained ResNet showed much worse performance vs. the pretrained SWIN-transformer (but still higher than the trained-from-scratch SWIN-transformer). That suggests transformer’s cross-attention between different time series (or different regions of the plot) might make a difference.
Should we use it? Probably not – lots of compute and memory is being wasted here producing mostly empty pixels. But it’s a sign of the coming trend of leverage pre-trained model or die trying.
Training with heterogenous multi-modal data
Data collection sucks and everyone knows it, especially neuroscientists. How we wish we can just bust out some kind of Imagenet, CIFAR, or COCO like the vision people? Nope, datasets are always too heterogenous in sensors, protocols, or modalities. Transformers are now making it easier to combine them now though (see for example previous).
BIOT: Biosignal Transformer for Cross-data Learning in the Wild
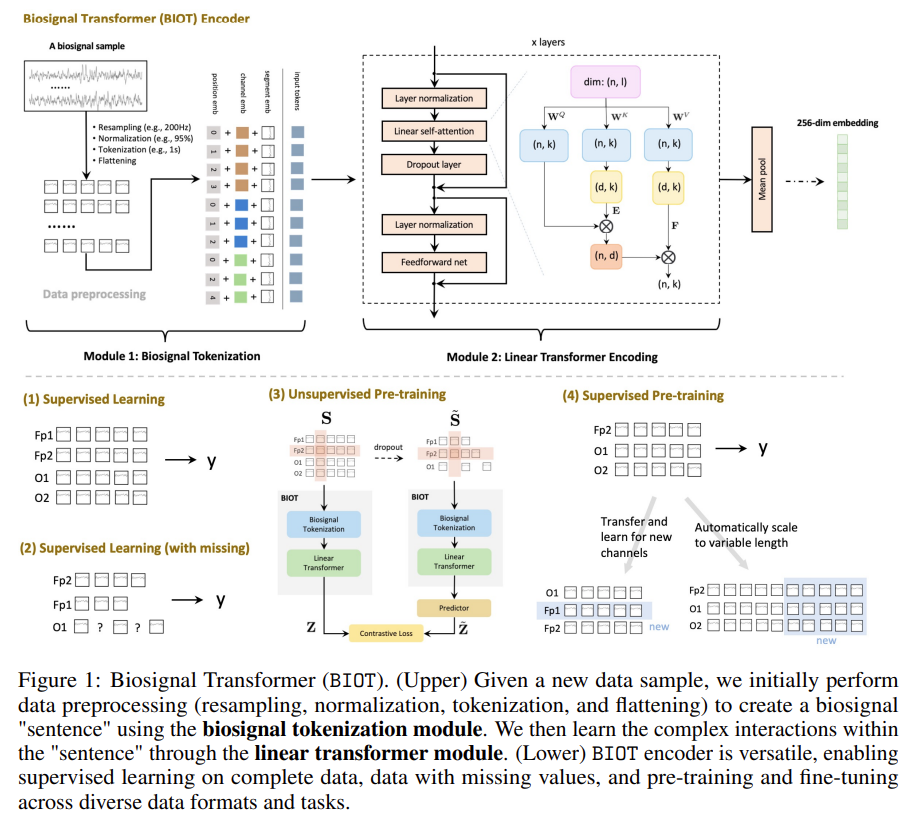
Main contributions:
- Biosignal transformer (BIOT): a generic biosignal learning model BIOT by tokenizing biosignals of various formats into unified “sentences.” •
- Knowledge transfer across different data: BIOT can enable joint (pre-)training and knowledge transfer across different biosignal datasets in the wild, which could inspire the research of large foundation models for biosignals.
- Strong empirical performance. We evaluate our BIOT on several unsupervised and supervised EEG, ECG, and human sensory datasets. Results show that BIOT outperforms baseline models and can utilize the models pre-trained on similar data of other formats to benefit the current task.
Fancy word aside, the main takeways:
- Segment time series into 1s chunks (called tokens). Then parametrize with 3 embeddings: [channels, samples] –> [(dim_emb1 + dim_emb2 + dim_emb3),].
- Pass them through a linear transformer (use reduced-rank form of self-attention).
- Profit with transformer embedding outputs..
This is very similar approach to Poyo1, which uses relative position embedding and does not need to explicitly chunk 1s windows.

Interestingly, BIOT paper claims to be “the first multi-channel time series learning model that can handle biosignals of various formats”. And both BIOT and POYO1 are in the Neurips 2023.
Leveraging LLM for decoding
Continuing onto the trend of using LLM at the end of all the ML stacks.. such as decoding mental image by conditioning diffusion models on fMRI, now we can decode “language” from EEG much better.
DeWave: Discrete Encoding of EEG Waves for EEG to Text Translation
Context:
Press releases such as this one would have you believe they have “developed a portable, non-invasive system that can decode silent thoughts and turn them into text”.
But what exactly are the “silent thoughts”?
study participants silently read passages of text while wearing a cap that recorded electrical brain activity through their scalp using an electroencephalogram (EEG)
This is different from what we typically think of “thoughts”, it’s more similar to decoding movies from neural activities (similar to Alexander Huth and Joe Culver works). Now we continue:
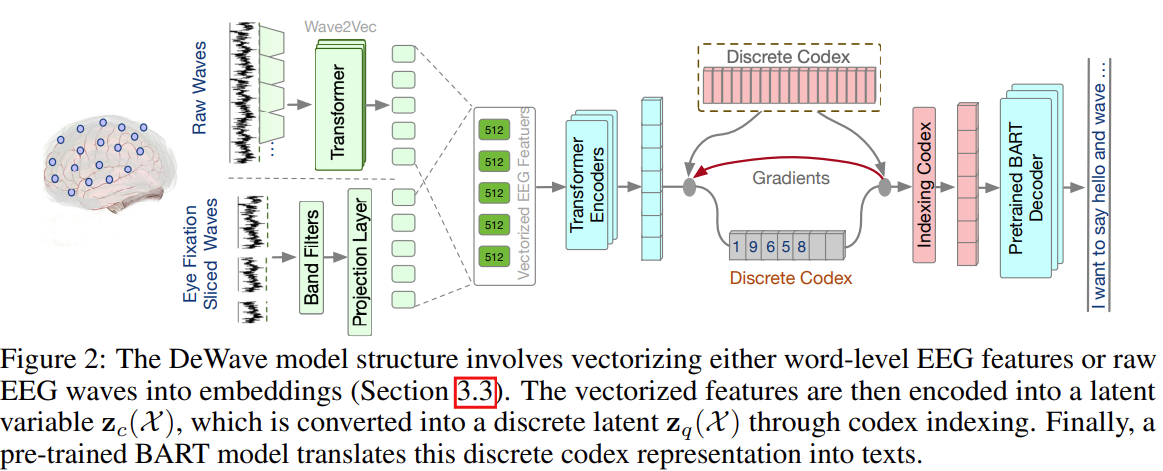
Main contributions:
- This paper introduces discrete codex encoding to EEG waves and proposes a new framework, DeWave, for open vocabulary EEG-to-Text translation.
- By utilizing discrete codex, DeWave is the first work to realize the raw EEG wave-to-text translation, where a self-supervised wave encoding model and contrastive learning-based EEG-to-text alignment are introduced to improve the coding ability.
- Experimental results suggest the DeWave reaches SOTA performance on EEG translation, where it achieves 41.35 BLUE-1 and 33.71 Rouge-1, which outperforms the previous baselines by 3.06% and 6.34% respectively
The paper does decoding with/without training data markers indicating where the subject’s looking at. The case without markers is much more interesting and sidesteps the labeling problem.
The overall approach:
- Use a conformer to vectorize the EEG signals into embeddings,
- the embeddings are mapped to a set of discrete “symbols” (or code) via a learned “codex”,
- The codex representations are fed into pre-trained BART (BERT+GPT) and get the output hidden states. A fully connected layer is applied on the hidden states to generate English tokens from pre-trained BART vocabulary V.
It’s hard to decipher some of the details of this paper, but recording notes here for future me.
Training paradigm:
- In the first stage, they do not involve the language model in weight updates. The target of the first stage is to train a proper encoder projection to theta_codex and a discrete codex representation C for the language model.
- In the second stage, the gradient of all weights, including language model theta_BART is opened to fine-tune the whole system.
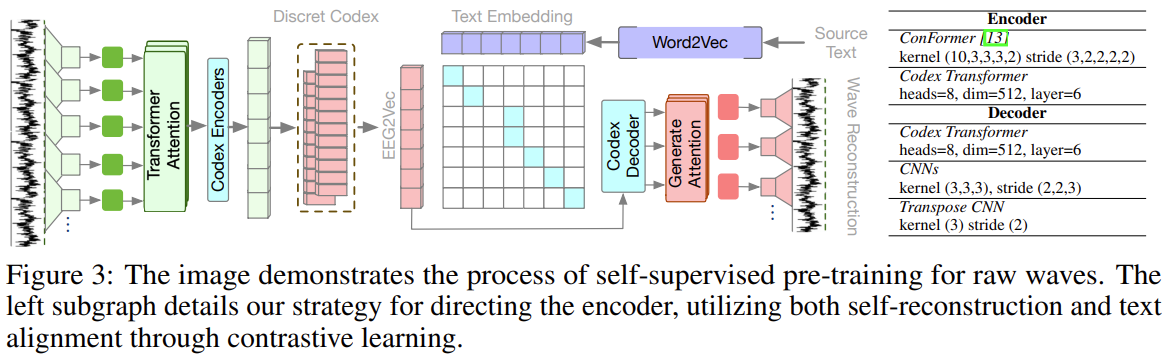
The codex approach is very interesting – instead of feeding EEG embeddings directly to the pre-trained BART, it gets converted into this intermediate representation. The rationale given was this:
It is widely accepted that EEG features have a strong data distribution variance across different human subjects. Meanwhile, the datasets can only have samples from a few human subjects due to the expense of data collection. This severely weakened the generalized ability of EEG-based deep learning models. By introducing discrete encoding, we could alleviate the input variance to a large degree as the encoding is based on checking the nearest neighbor in the codex book. The codex contains fewer time-wise properties which could alleviate the order mismatch between event markers (eye fixations) and language outputs.
Need temporal alignment between segments of signals and word (or “label”?).
Not sure if this is back-rationalization, but the training supports it:
To train the encoder as well as the codex, two self-supervised approach was used:
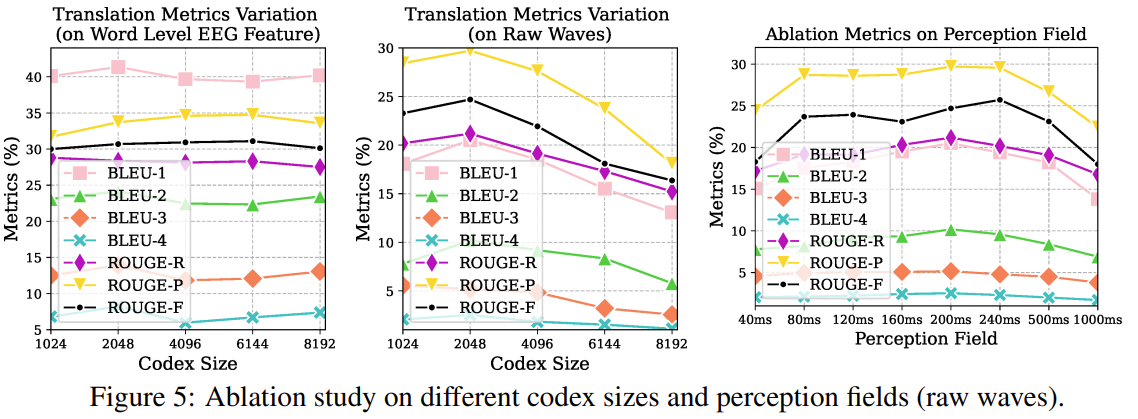
- Encoder-decoder Reconstruction: raw waves -> embeddings -> codex -> embeddings -> raw waves. Notably subsequent ablation studies showed that a larger codex size is not necessarily better, which makes sense here as we expect a lower-D latent space for this approach to work.
- Language (word2vec embeddings) and codex alignment via contrastive learning (of course!)
And this approach works, even though EEG is traditionally very shitty. Sure they used new graphene-based dry electrodes which supposedly approach wet-gel electrode performance, it’s still surprising. Though I’m not versed in EEG to understand how significant the outperformance margin vs. SOTA is.
I don’t buy the jusification for the codex representation though, as the abolation study shows the effect codex on word level EEG feature as minimal.
Predicting brain responses with large pre-trained models
Alex Huth is making it rain in Neurips this year with a series of fMRI response encoding papers, involving language and vision/video. One that stood out the most to me was Scaling laws for language encoding models in fMRI, also see tweet-thread.
Takeaways:
- Predicting fMRI brain response to story-listening with LLM: Brain prediction performance scales logarithmically with model size from 125M to 30B parameter models, with ~15% increased encoding performance as measured by correlation with a held-out test set across 3 subjects. Similar logarithmic behavior was observed when scaling the size of the fMRI training set.
- Similar trend exists for acoustic encoding models that use HuBERT, WavLM, and Whisper.
A noise ceiling analysis of these large, high-performance encoding models showed that performance is nearing the theoretical maximum for brain areas such as the precuneus and higher auditory cortex. These results suggest that increasing scale in both models and data will yield incredibly effective models of language processing in the brain, enabling better scientific understanding as well as applications such as decoding.
On the surprising effectiveness of neural decoding with large pre-trained models..
Deep-learning models were only “weakly” inspired by the brain and even though the Transformer arch isn’t exactly neuro-inspired, decoding and neural responses using large pre-trained models have been surprisingly effective and “straight-forward”.
My take is that these large pre-trained models (language, vision, audio, etc) encode structure of that particular modality. Brains proces these modalities differently from these models, but presumably there’s some kind of latent space that both can be mapped onto. So this stype of encoder-decoder approach can be thought of as latent space alignment (which can include temporal dimension as well!)
This is a step up from other dynamic-programming style alignment techniques such as viterbi decoding and CTC-loss, and much more elegant conceptually.
